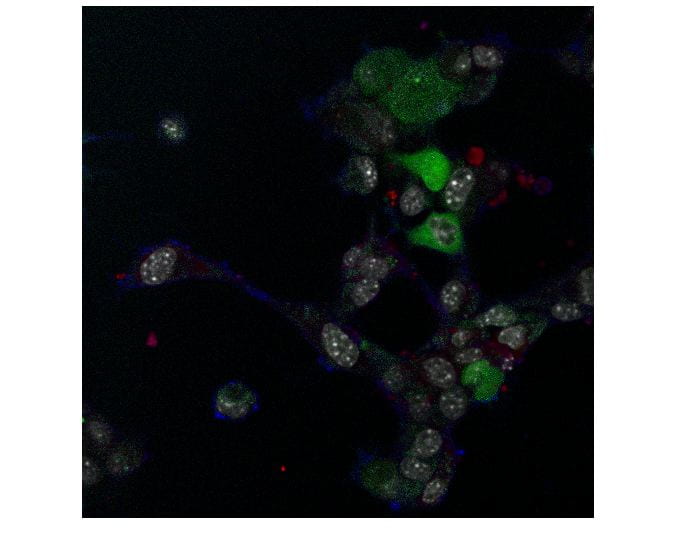
In the last decade, many different tools have been invented for evaluation of protein-protein interactions, which is important for understanding different diseases such as pneumonia. As part of big research on cell interactions, video sequences of hyperspectral images of the cells are taken and analyzed. The problem is that there is a huge amount of cells and images, and thus it is very hard to use manual segmentation and tracking.
In this project, we use hyperspectral images of cells to find their boundaries, their nuclei, and to identify the motion within the cell. This is not an easy task since the boundaries are not well defined. In addition, we gather statistical data on the cells and measure FRET efficiency.

Published by