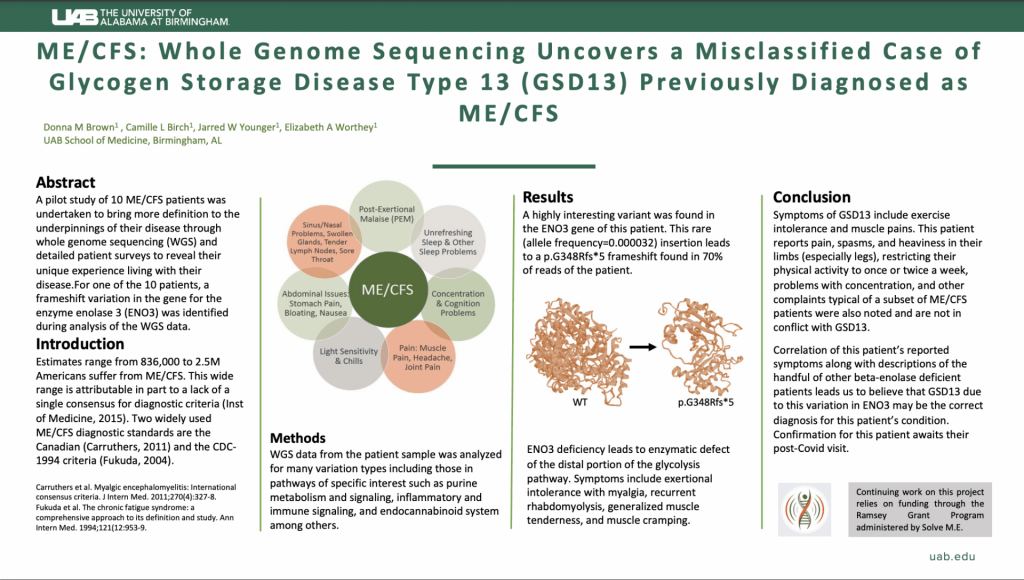
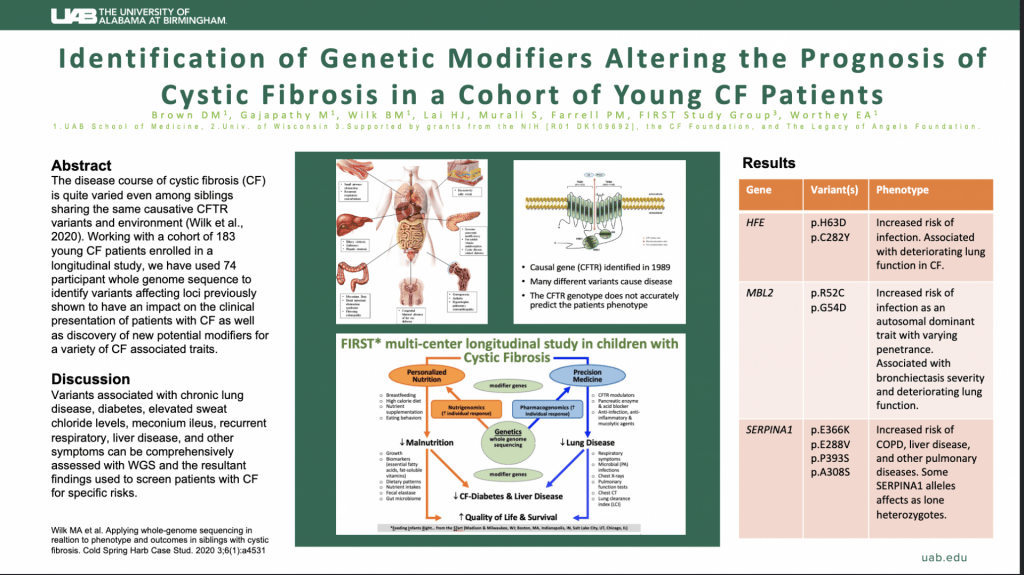
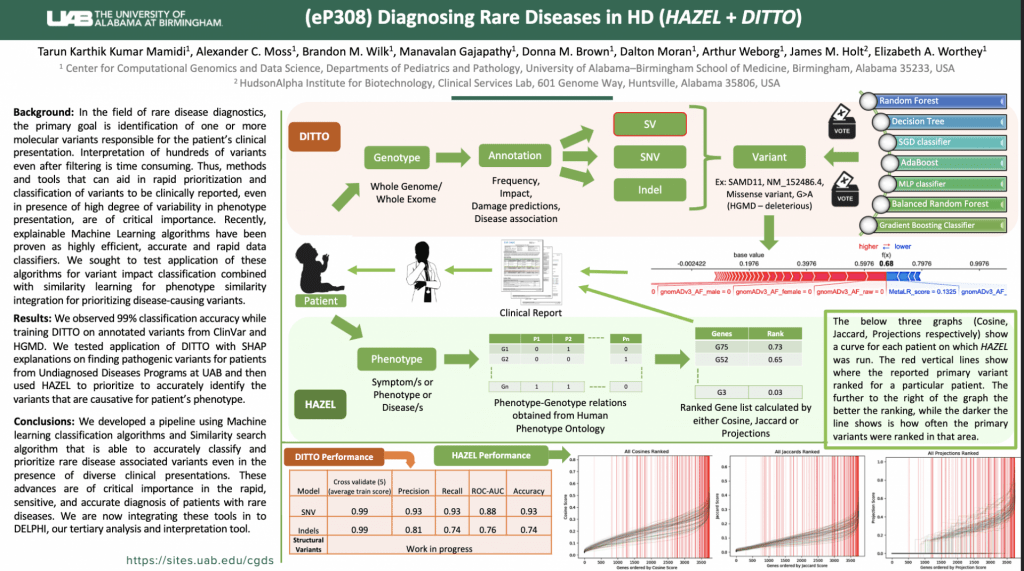
Apply Here: https://uab.taleo.net/careersection/ext/jobdetail.ftl?job=T186075&tz=GMT-06%3A00&tzname=
Come join a diverse and growing team of computational biologists working to develop and apply computational biology, bioinformatics, and data science methods to identify the molecular underpinnings of disease and to generate knowledge of the functional impact of molecular variation to better understand disease onset, progression, and outcome, response to therapeutics, and relationship to human health. This individual will have the opportunity to work with world class researchers and clinicians and to work on a variety of pediatric and adult diseases. Current projects are focused on gaining knowledge and understanding of disease mechanisms and outcomes for Mendelian disorders and more common complex diseases including cancer, neurodegenerative diseases, and immune disorders.
Mission, Vision & Values
UAB serves students, patients, the community and the global need for discovery, knowledge dissemination, education, creativity and the application of groundbreaking solutions. We are a leader among comprehensive public urban research universities with academic medical centers. We expect all employees to adhere to our shared values of Integrity, Respect, Diversity and inclusiveness, Collaboration, Excellence and achievement, Stewardship, and Accountability. UAB’s Vision, Mission & Shared Values can be found here: https://www.uab.edu/plan/the-plan/mission-vision-shared-values/
General Responsibilities
To participate in research activities involved in the analysis of Next Generation Sequencing such as data derived from genomics and transcriptomics (bulk and single-cell sequencing) as part of the UAB Biological Data Science institutional core. To support research services such as study design, data management, data analysis, pipeline development and to aid in the generation of training guides and documentation. To stay abreast of recent literature and other new developments, incorporating new techniques into existing data analysis protocols and pipelines in support of the institutional core’s mission of high quality, reproducible computational biology research.
Key Duties & Responsibilities
1. Performs computational data analysis of transcriptomics and genomics studies.
2. Designs, develops and implements software tools and analytical procedures supporting bioinformatics analysis.
3. Participates in projects related to the core sustainability including but not limited to pipeline development, web application development, maintenance of documentation and training guides and other services to support the needs of UAB investigators.
4. Provides computational support services such as study design, data management and datamigration.
5. Offers bioinformatics consultation services to UAB investigators.
6. Evaluates and identifies new technologies for implementation.
7. Performs other duties as assigned.
Minimum Requirements
Doctorate in a related field OR Master’s degree in a related field and two (2) years of related experience OR Bachelor’s degree in related field and four (4) years of related experience OR Associate’s degree in related field and six (6) years of related experience required. Work experience may NOT substitute for education requirements.
Knowledge/Skills/Abilities
●Knowledge of the biological sciences
●Experience in computational biology analysis
●Experience in Linux and using a command-line environment
●Knowledge of scripting languages such as, Python, shell scripts, and R
●Knowledge of best practices of reproducible and scalable research such as version control (git) and environment managing systems (e.g. Conda, Docker, Singularity)
●Proficiency in the visualization of omics data
●Experience in High Performance Computing environment is preferable
●Ability to work as part of a team
●Knowledge of theoretical and applied statistics, and molecular biology, as demonstrated through previous work or academic experience
●Problem-solving skills
●Attentive to detail
●Organizational skills
●Excellent written and verbal communication skills
An ideal candidate will follow best practices described to ensure reproducible and scalable research and be proficient in the analysis of transcriptomics data (e.g.: differential expression analysis, enrichment analysis, single-cell or nuclei data visualization and analysis, gene set annotation and contribute to upstream data analysis through the development of pipelines). An ideal candidate should also be familiar with managing virtual computational environments and be familiar with high performance computing clusters.DisclaimerPlease Note: The duties and responsibilities described are not a comprehensive list and additional tasks may be assigned to the employee as necessitated by business demands. This job description does not constitute a contract of employment or otherwise limit UAB’s employment-at-will rights at any time. Employees are expected to comply with all UAB policies and procedures during their employment.
Disclaimer
Please Note: The duties and responsibilities described are not a comprehensive list and additional tasks may be assigned to the employee as necessitated by business demands. This job description does not constitute a contract of employment or otherwise limit UAB’s employment-at-will rights at any time. Employees are expected to comply with all UAB policies and procedures during their employment